Lots of interesting abstracts and cases were submitted for TCTAP 2023. Below are the accepted ones after a thorough review by our official reviewers. Don’t miss the opportunity to expand your knowledge and interact with authors as well as virtual participants by sharing your opinion in the comment section!
TCTAP A-078
Identification of Underlying Genes and Pathways Associated With Myocardial Ischemia-Reperfusion Injury via Bioinformatic Analysis
By Jing Xu
Presenter
Jing Xu
Authors
Jing Xu1
Affiliation
Fuwai Hospital, China1
View Study Report
TCTAP A-078
Genomics/Proteomics
Identification of Underlying Genes and Pathways Associated With Myocardial Ischemia-Reperfusion Injury via Bioinformatic Analysis
Jing Xu1
Fuwai Hospital, China1
Background
With the development of percutaneous coronary, anti-platelet drugs, anti-thrombotic drugs and cardiac surgery, myocardial ischemia-reperfusion injury (MIRI) has received more and more attention. Myocardial ischemia-reperfusion injury (MIRI) is the recovery of blood perfusion in myocardial tissue after a long period of ischemia, which can cause more serious and more obvious damage and dysfunction reperfusion, including decreased systolic function, decreased coronary flow and vascular reactive changes. It can be seen that myocardial ischemia-reperfusion injury has become a major obstacle in clinical treatment, and its harm cannot be ignored. Studying its pathogenesis and exploring effective prevention and treatment measures are of great significance for the treatment of myocardial infarction. Our objectives were i) to determine the underlying genes and molecular pathways associated with MIRI via bioinformatic analysis and (ii) to identify drugs targeting the relevant MIRI molecular pathways.
Methods
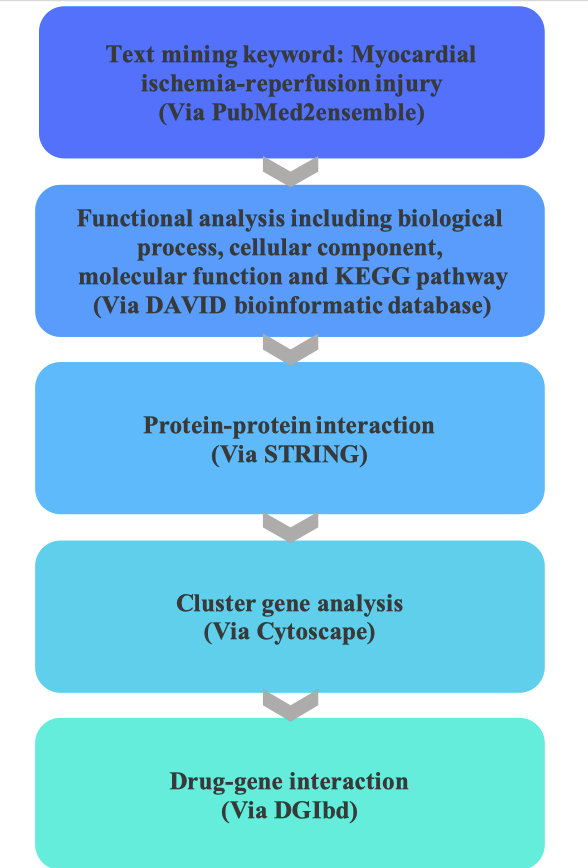
MIRI-associated genes were searched by text mining, and the intersection of the two gene sets was selected for gene ontology analysis via DAVID database. Protein interaction network analysis was performed via STRING. Enriched cluster genes were analyzed by Cytoscape software. Drug-gene interactions were performed by DGIdb.

Results
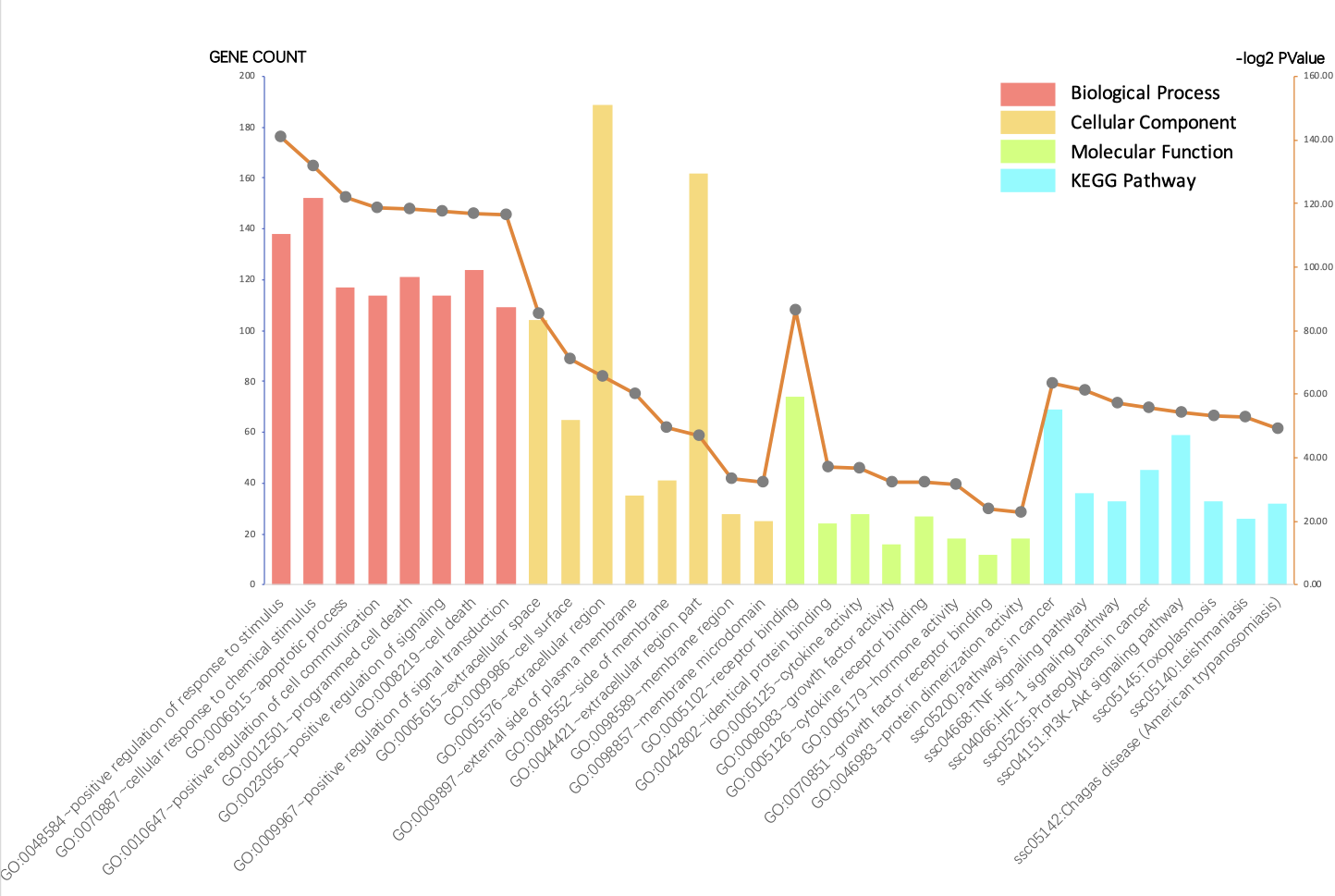
552 genes associated with MIRI were identified. A gene cluster including CXCL10, POMC, CXCR3, CX3CR1, CCR5, CCL5, ADCY3, CNR1, ANXA1, C5, FPR2, C5AR1, ADCY7, CCL4, AGTR2, FPR3, KNG1, CNR2 and AGT, coupled with chemokine signaling pathway was found to be significant. 14 candidate drugs were targeted as possible mediators in the process of MIRI. According to further analysis, ANXA1 would be a regulator in MIRI affecting the process of programmed cell death. ADCY7, ADCY3 and CX3CR1 were also found underlying mechanisms for cardioprotective function in MIRI.

Conclusion
This study has improved the comprehensive understanding of the pathogenesis and underlying molecular mechanism in MIRI and proved the availability of bioinformatic methods to search the novel possible connections between genes and diseases. These selected candidate genes and pathways could show us new clues to search the therapeutic targets for the treatment of MIRI. What’s more, this study could also provide the ideas for searching the mechanisms of effective traditional Chinese medicines for MIRI. However, further studies including molecular biological experiments are required to confirm the function of these identified genes in MIRI.




