Lots of interesting abstracts and cases were submitted for TCTAP 2023. Below are the accepted ones after a thorough review by our official reviewers. Don’t miss the opportunity to expand your knowledge and interact with authors as well as virtual participants by sharing your opinion in the comment section!
TCTAP A-077
Genetic Variants of TTN and DMD Highly Correlated With Existence of Severe Myocardial Bridging
By Tsung Lin Yang, Chun-Ming Shih
Presenter
Tsung Lin Yang
Authors
Tsung Lin Yang1, Chun-Ming Shih1
Affiliation
Taipei Medical University Hospital, Taiwan1
View Study Report
TCTAP A-077
Genomics/Proteomics
Genetic Variants of TTN and DMD Highly Correlated With Existence of Severe Myocardial Bridging
Tsung Lin Yang1, Chun-Ming Shih1
Taipei Medical University Hospital, Taiwan1
Background
Myocardial bridge (MB) is a congenital coronary artery anomaly and an important cause of chest pain. The genetic basis of this cardiac vasculopathy is unknown.
Methods
Whole-exomes sequencing technique was exploited to collect genetic information from MB and normal subjects. Genotypic differences were analyzed with GATK pipelines. Functional annotations were investigated via Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, and knockout mouse database. Pathogenicity prediction was performed by ClinVar, CADD, and REVEL scoring systems.
Results
From April 2020 to May 2021, 248 subjects who had undergone coronary angiographies were screened, and MBs were detected in 46 (18.5%) patients. MB severity was ranked based on the luminal area loss during systole. Eight pairs of angiographically-identified severe myocardial bridge patients and normal controls were enrolled for genetic analysis. One hundred and fifty-three candidate variants in 140 genes potentially pathogenic for MB were identified. Functional annotation of candidate genes indicated a significant association with abnormal skeletal muscle mass, cardiomyopathy, and transmembrane cation proteins. Genes with the most variants ranked from the ClinVar and CADD/REVEL were the TTN, DMD, and TCF7L1, SOX8, respectively. Combining the number of variants, existence in MB patients, and tissue expressions, TTN and DMD gene polymorphisms were the most significantly associated with the existence of a myocardial bridge. The TCF7L1 and SOX8 genes contributed to multiple loci of pathogenic variants, which collectively predicted 25% of MB. Gene variants on the 4 genes (TTN, DMD, TCF7L1, and SOX8) predicted MB with a sensitivity of 87.5% and a specificity of 100%.
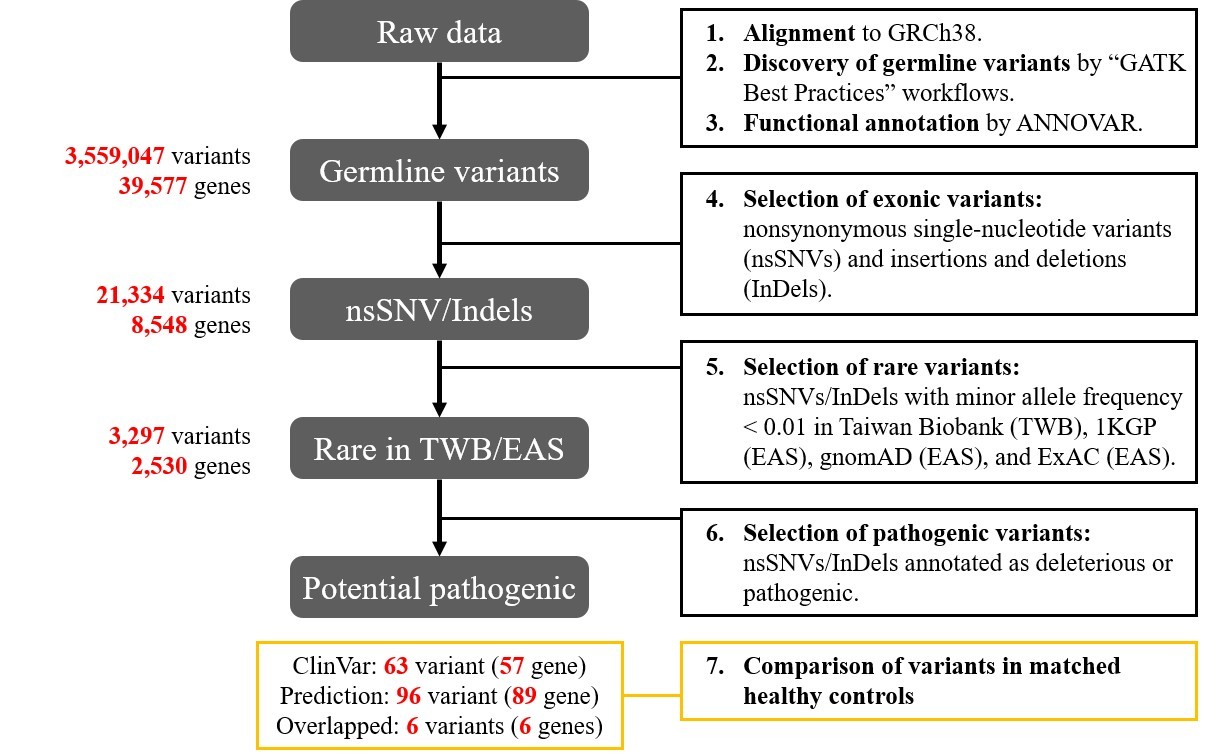
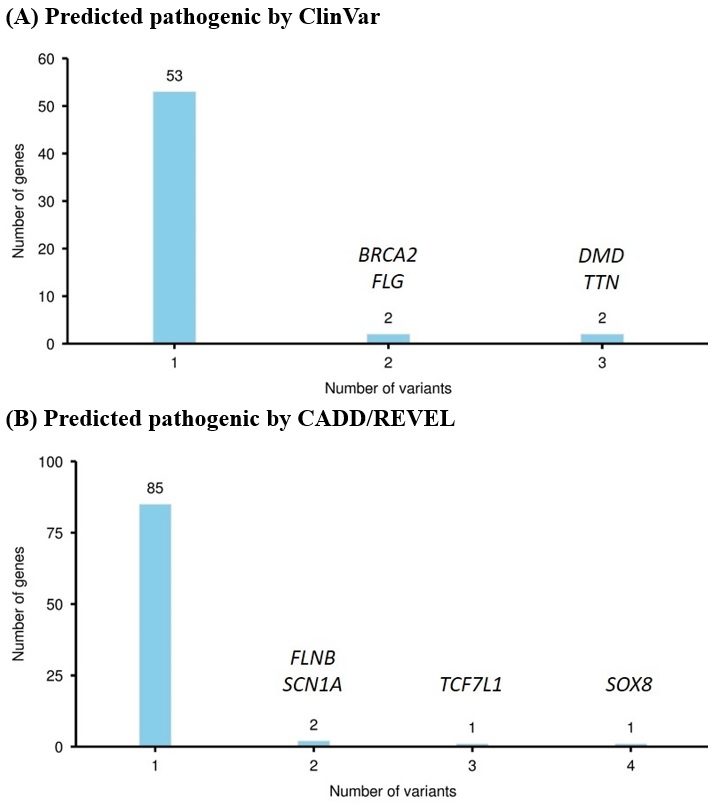
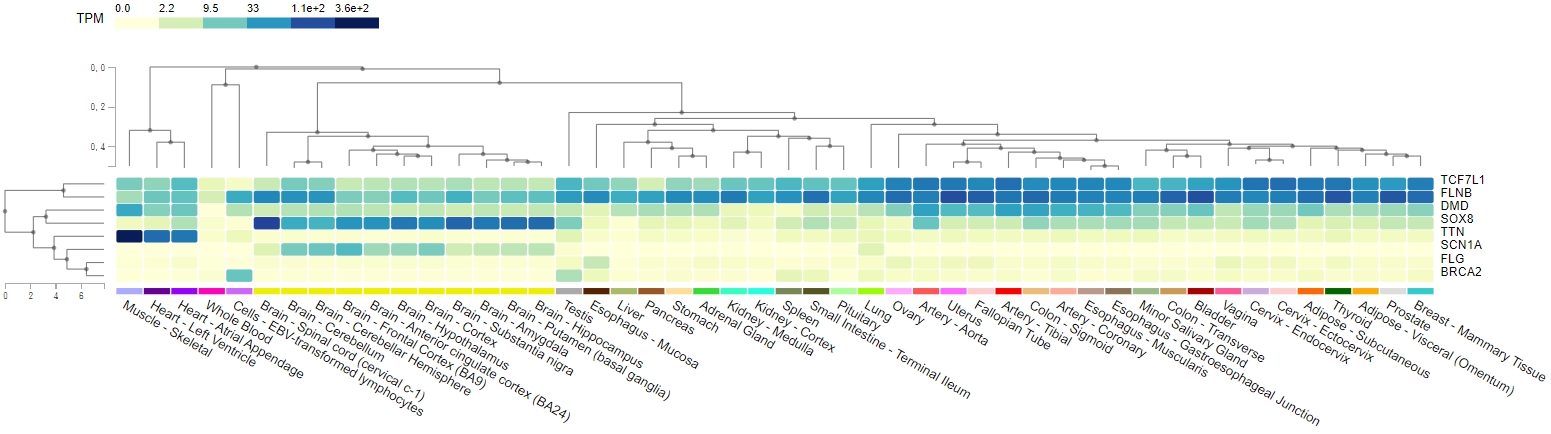



Conclusion
Genetic variants on TTN, DMD, TCF7L1, and SOX8 were highly associated with the myocardial bridge, especially the first two genes. Further studies would be needed to clarify the interplays between cardiac sarcomeric protein titin and cardiomyocyte anchoring protein dystrophin in the formation of the myocardial bridge.


