Lots of interesting abstracts and cases were submitted for TCTAP 2022. Below are the accepted ones after a thorough review by our official reviewers. Don’t miss the opportunity to expand your knowledge and interact with authors as well as virtual participants by sharing your opinion in the comment section!
TCTAP A-007
Circulating Cell-Free DNA 5hmC Modifications as Candidate Biomarkers for ST-Segment Elevated Myocardial Infarction
By Jing Xu
Presenter
Jing Xu
Authors
Jing Xu1
Affiliation
Fuwai Hospital, China1
View Study Report
TCTAP A-007
Acute Coronary Syndromes (STEMI, NSTE-ACS)
Circulating Cell-Free DNA 5hmC Modifications as Candidate Biomarkers for ST-Segment Elevated Myocardial Infarction
Jing Xu1
Fuwai Hospital, China1
Background
Acute myocardial infarction (AMI) remains a common cardiac emergency incidence worldwide. The inability to accurately and temporally predict the occurrence of AMI impairs our capability to further improve patient outcomes. Moreover, survivors of acute ST-segment elevated myocardial infarction (STEMI) are prone to develop progressive ventricular remodeling and dysfunction that leads to heart failure (HF). DNA 5-hydroxymethylcytosine (5hmC) modification is an epigenetic marker involved in a range of biological processes. Besides, accumulating evidence suggest that 5hmC and its TET2 enzyme, one member of the TET family, play an important role in atherosclerosis and are not only involved in the regulation of vascular smooth muscle cell phenotype, but also closely related to endothelial dysfunction and inflammatory immune response. However, little information is available about its role in coronary artery disease (CAD), particularly in the acute phase of STEMI.
Methods
We utilized 5hmC-Seal to generate genome-wide 5hmC profiles in plasma cell-free DNA (cfDNA) of normal coronary artery (NCA, n = 182) controls and STEMI patients (n = 193). Characteristics of 5hmC distribution and GO enrichment of differentially expressed genes were explored. To investigate the correlation between 5hmC modifications and STEMI, we separated samples into training, testing and validation cohorts and developed a 5hmC-based logistic regression model from the training cohort to predict the progression of STEMI.
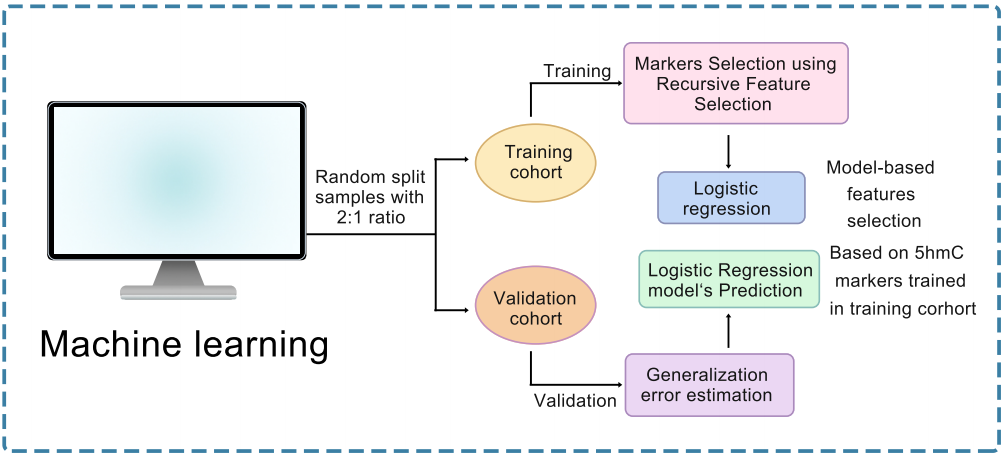
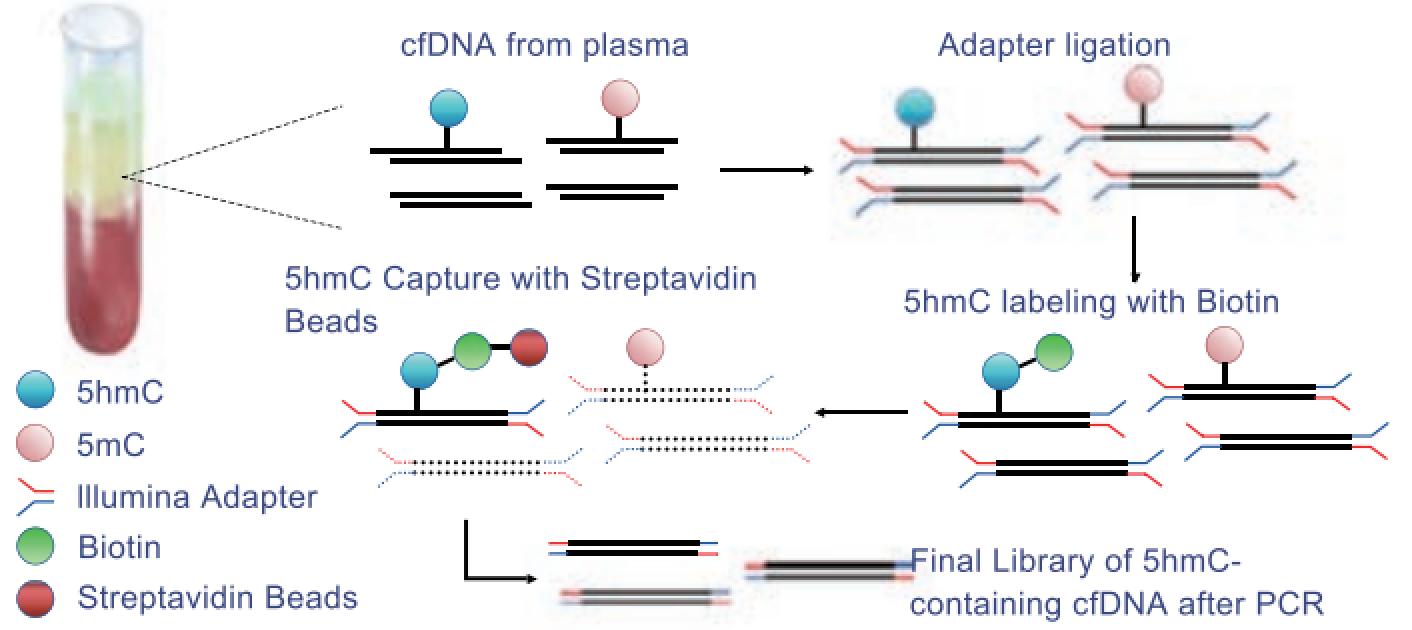


Results
We detected a significant difference of 5hmC enrichment in gene bodies from STEMI patients compared with NCA individuals including DOCK4, MAP4K4, HDAC4, PDE4D, PDE8B, and BCL6. Particularly, our results showed that patients of STEMI can be well separated from NCA individuals by 5hmC markers. The prediction performance of the model established by differentially regulated 5hmC modified genes achieved an AUC of 0.78 and 0.74 in validation cohorts.
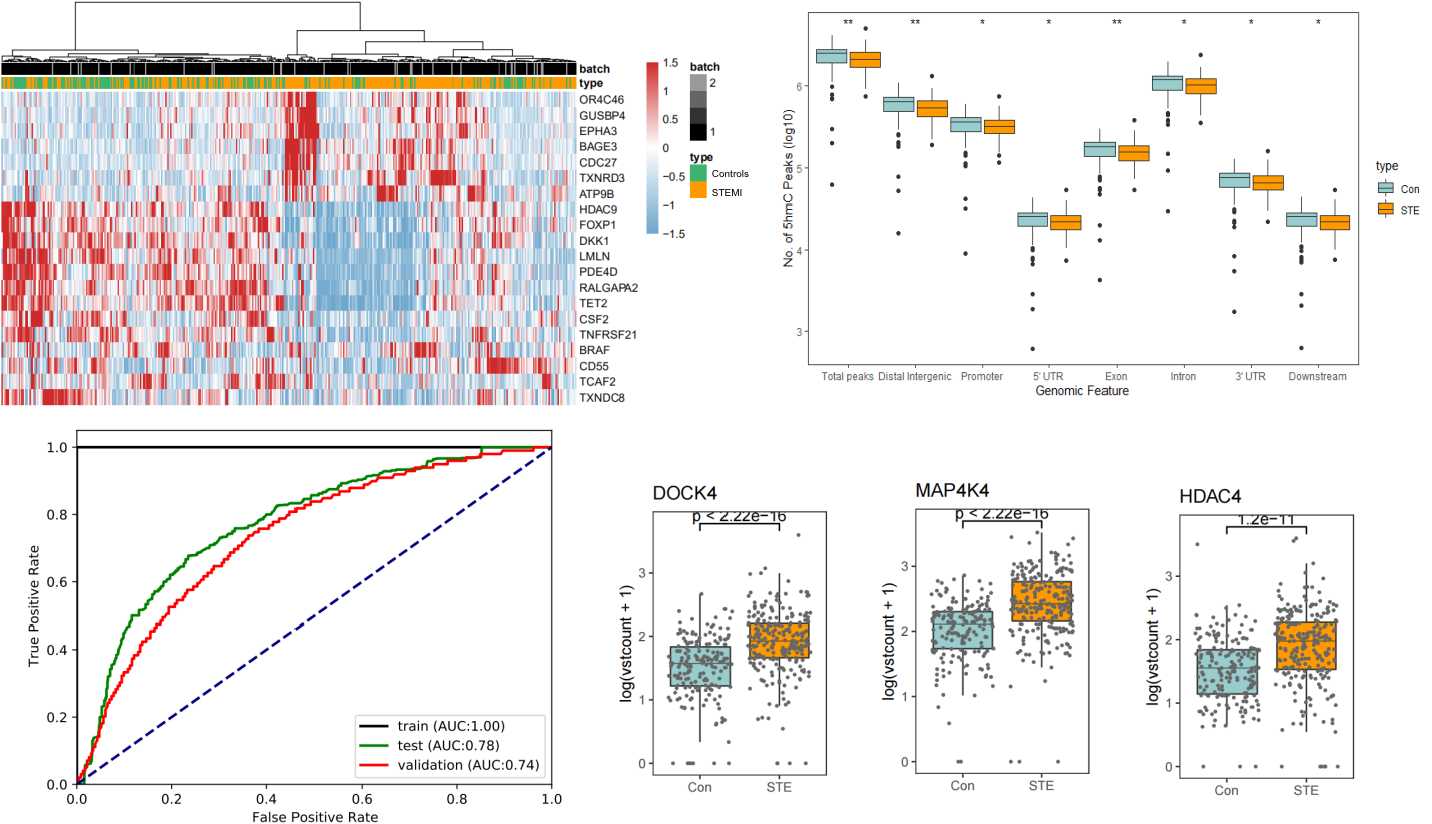

Conclusion
Our results demonstrated that patients of STEMI and NCA individuals had distinct differences in 5hmC enrichment. 5hmC markers derived from plasma cfDNA may potentially serve as a clinical-applicable, minimally invasive, and liquid biopsy-based approach to diagnose CAD, particularly used to predict the occurrence of fetal STEMI.


