Lots of interesting abstracts and cases were submitted for TCTAP 2025. Below are the accepted ones after a thorough review by our official reviewers. Don’t miss the opportunity to expand your knowledge and interact with authors as well as virtual participants by sharing your opinion in the comment section!
TCTAP A-097
The In-Silico Analysis of Andrographolide and Its Derivatives to Mitigate Hypertensive-Induced Cardiac Remodeling: Insights From Differential Gene Expression and Computational Modeling
By Muhamad Rizqy Fadhillah, Wawaimuli Arozal, Heri Wibowo, Aryo Tedjo, Suci Widya Primadhani, Nurul Gusti Khatimah, Clara Riski Amanda
Presenter
Muhamad Rizqy Fadhillah
Authors
Muhamad Rizqy Fadhillah1, Wawaimuli Arozal1, Heri Wibowo1, Aryo Tedjo1, Suci Widya Primadhani2, Nurul Gusti Khatimah1, Clara Riski Amanda1
Affiliation
Universitas Indonesia, Indonesia1, Cileungsi Public Regional Hospital, Indonesia2
View Study Report
TCTAP A-097
Digital Health and Artificial Intelligence
The In-Silico Analysis of Andrographolide and Its Derivatives to Mitigate Hypertensive-Induced Cardiac Remodeling: Insights From Differential Gene Expression and Computational Modeling
Muhamad Rizqy Fadhillah1, Wawaimuli Arozal1, Heri Wibowo1, Aryo Tedjo1, Suci Widya Primadhani2, Nurul Gusti Khatimah1, Clara Riski Amanda1
Universitas Indonesia, Indonesia1, Cileungsi Public Regional Hospital, Indonesia2
Background
The overload pressure caused by systemic hypertension is a well-recognized initiator of cardiac remodeling. While clinically approved anti-cardiac remodeling drugs exist, the quest for safer and more effective compounds persists. Natural products have historically provided a rich source of bioactive metabolites, frequently evaluated for their potential as drug candidates. Andrographis paniculata, a plant rich in andrographolide and native to Indonesia, has shown promise for its anti-cardiac remodeling properties. Our research group’s in vitro and in vivo experiments demonstrated that both crude extracts of Andrographis paniculata and andrographolide exhibited cardioprotective effects in cardiac hypertrophy and toxicity models. However, optimizing the crude extract and its isolated compounds is essential to enhance their efficacy and pharmacokinetic characteristics. Andrographolide, a diterpene lactone, is well-documented for its anti-cardiac remodeling effects. Despite this, traditional drug discovery processes for compound optimization can be time-consuming, often taking up to adecade to achieve clinical approval. Computer-aided drug design (CADD) has revolutionized the drug discovery landscape by efficiently predicting the lipophilicity, bioactivity, and binding modes of molecules with specific drug targets. Considering this, we conducted a study to evaluate the potential of andrographolide and its derivatives, employing molecular docking and molecularfinger print analysis to predict bioactivity and lipophilicity metrics. Differentially expressed genes (DEGs) analysis using publicly available transcriptomic data was performed to identify significant pathways and druggable protein targets associated with these pathways.
Methods
The DEGs analysis was conducted using a publicly available dataset from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/)with the GEO2R tool. A dataset of spontaneous hypertensive rat models was selected to mimic pressure overload-induced cardiac remodeling. The data weredivided into normal and diseased rat groups and validated using principal component analysis (PCA) with Orange® software. For exploratory analysis, afternormalization, the significance level was adjusted using the Benjamini and Hochberg method, with a threshold set at padj < 0.1 and a Log2-Fold Change threshold of 1.
Results
The dataset GSE2116 was selected to identify differentially expressed genes (DEGs) associated with cardiac remodeling induced by overload pressure at the tissue level. This dataset provides insights into the gene expression transition from hypertrophy to heart failure, observed on days 0, 12, 16, and 20. Principal Component Analysis (PCA) confirmed the differentiation between healthy and diseased rats. A total of 621,306, and 291 DEGs were identified for the comparisons of normal vs. day 12, normal vs. day 16, and normal vs. day 20, respectively, with 44 DEGs being shared between days 12 and 20. Our analysis indicated that the HIF-1α signaling pathway had the highest fold-enrichment on day 12, while the AGE-RAGE signaling pathway became increasingly prominent on days 16 and 20, suggesting it was the most significant contributor to cardiac remodeling progression. Therefore, the RAGE protein (PDB ID 6XQ5) was chosen as the druggable target for molecular docking and molecular fingerprint analysis. Andrographolide and its 29 derivatives were docked against the RAGE protein and displayed negative binding energies, indicating spontaneous ligand-receptor interactions. For molecular fingerprint analysis via machine learning, 73 compounds along with their IC50 values were extracted, and the data were split into 80% training and 20% testing sets. Notably, CID44593583 (Compound 8) with a MolDock score of-105.358 kJ/mol, CID45358107 (Compound 22) with -108.43 kJ/mol, andCID11968631 (Compound 28) with -108.97 kJ/mol exhibited superior binding energy compared to andrographolide (Compound 6) with -81.859 kJ/mol. These three compounds also have comparable MolDock Score with Azeliragon, a standard RAGE inhibitor (MolDock Score -117.42 kJ/mol). However, Compound 8, 22, and 28 also show comparable IC50 lipophilicity metrics profiles with Compound 6. Despite strong interactions with RAGE protein, these three compounds do not have good absorption profiles compared with Compound 6.
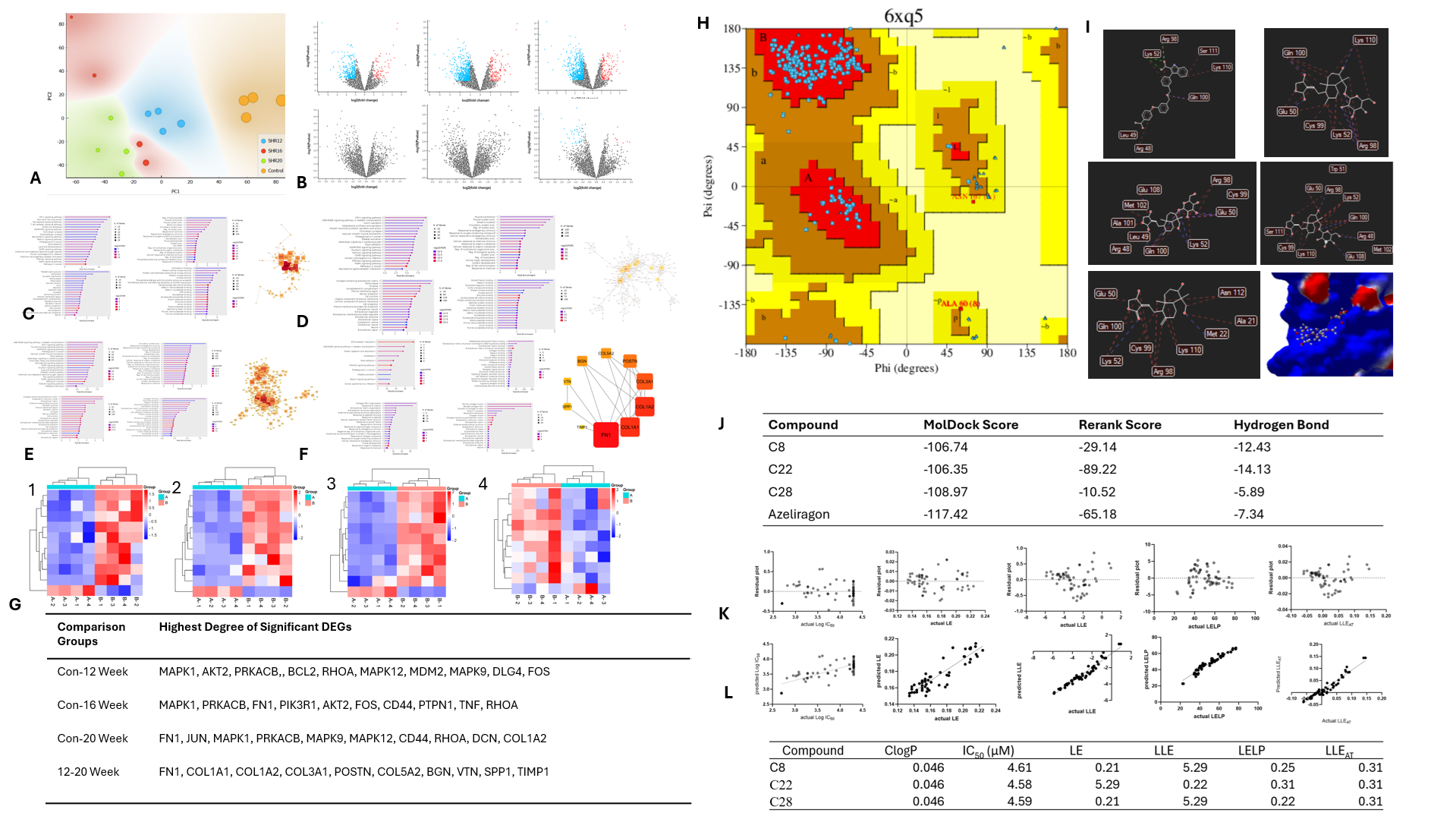

Conclusion
Our study reveals that the AGE-RAGEsignaling pathway contributes significantly to the development of cardiacremodeling. Furthermore, in-silico optimization identified that compound 8,compound 22, and compound 28 show superior binding activity with the RAGEactive sites compared to andrographolide and comparable with Azeliragon. Furthermore,these three compounds have favorable predicted bioactivity and ligandefficiency-lipophilicity metrics profiles. Despite their low absorptioncompared to andrographolide, these findings suggest that compound 8, 22, and28 derivatives may serve as promising drug candidates for inhibiting theRAGE signaling pathway to prevent cardiac remodeling progression, warrantingfurther preclinical verification and optimization for drug development.


